BACKRUB-LIKE BACKBONE SIMULATION RECAPITULATES NATURAL PROTEIN
CONFORMATIONAL VARIABILITY AND IMPROVES MUTANT SIDE-CHAIN PREDICTION
CONFORMATIONAL VARIABILITY AND IMPROVES MUTANT SIDE-CHAIN PREDICTION
Backrub-Like Backbone Simulation Recapitulates Natural Protein Conformational Variability And Improves Mutant Side-Chain Prediction
Smith, CA, Kortemme, T
J Mol Biol 380(4):742-56, 2008
Smith, CA, Kortemme, T
J Mol Biol 380(4):742-56, 2008
Proteins undergo conformational fluctuations in response to thermal energy, binding events, and mutation. Understanding and predicting such excursions around the native state of a protein is a key challenge in computational molecular biology. However, many applications keep the backbone structure fixed, while in actual proteins the backbone often undergoes subtle shifts in response to binding events or sequence changes. Successfully capturing such near-native shifts is thus important for many docking and design applications.
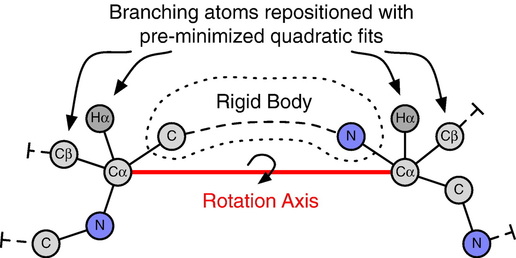
In this work, we developed a new local perturbation of protein backbones based on motions seen in high-resolution crystal structures. These fluctuations observed in the crystal lattice motivated Davis et al. to create a simple model, called “Backrub”, for subtle backbone shifts using just three residues. The core idea the work here is to use that type of motion, observed in nature, to computationally sample backbone configurations in a generalized scheme. We implemented a backrub-inspired sampling method in the Rosetta structure prediction and design program. We evaluated this model of backbone flexibility using three different tests: First, we show that Rosetta backrub simulations recapitulate the correlation between backbone and side-chain conformations in the high-resolution crystal structures upon which the model was based. Next, we show that backbone flexibility improves the accuracy of predicting point-mutant side-chain conformations over fixed backbone rotameric sampling alone. Finally, we show that backrub sampling of triosephosphate isomerase loop 6 can capture the millisecond/microsecond oscillation between the open and closed states observed in solution. Our results suggest that backrub sampling captures a sizable fraction of localized conformational changes that occur in natural proteins.
In this work, we developed a new local perturbation of protein backbones based on motions seen in high-resolution crystal structures. These fluctuations observed in the crystal lattice motivated Davis et al. to create a simple model, called “Backrub”, for subtle backbone shifts using just three residues. The core idea the work here is to use that type of motion, observed in nature, to computationally sample backbone configurations in a generalized scheme. We implemented a backrub-inspired sampling method in the Rosetta structure prediction and design program. We evaluated this model of backbone flexibility using three different tests: First, we show that Rosetta backrub simulations recapitulate the correlation between backbone and side-chain conformations in the high-resolution crystal structures upon which the model was based. Next, we show that backbone flexibility improves the accuracy of predicting point-mutant side-chain conformations over fixed backbone rotameric sampling alone. Finally, we show that backrub sampling of triosephosphate isomerase loop 6 can capture the millisecond/microsecond oscillation between the open and closed states observed in solution. Our results suggest that backrub sampling captures a sizable fraction of localized conformational changes that occur in natural proteins.